# Parametric survival models
---
{{< include shared-config.qmd >}}
## Parametric Survival Models
### Exponential Distribution
- The exponential distribution is the basic distribution for survival
analysis.
$$
\begin{aligned}
f(t) &= \lambda e^{-\lambda t}\\
\log{f(t)} &= \log{\lambda}-\lambda t\\
F(t) &= 1-e^{-\lambda t}\\
\surv(t)&= e^{-\lambda t}\\
\cuhaz(t) &= -\log{\surv(t)}
\\ &= \lambda t\\
\haz(t) &= \lambda\\
\text{E}(T) &= \lambda^{-1}
\end{aligned}
$$
### Weibull Distribution
Using the Kalbfleisch and Prentice (2002) notation:
$$
\begin{aligned}
f(t)&= \lambda p (\lambda t)^{p-1}e^{-(\lambda t)^p}\\
F(t)&=1 - e^{-(\lambda t)^p}\\
\surv(t)&=e^{-(\lambda t)^p}\\
\haz(t)&=\lambda p (\lambda t)^{p-1}\\
\cuhaz(t)&=(\lambda t)^p\\
\log{\cuhaz(t)} &= p \log{\lambda t}
\\ &= p \log{\lambda} + p \log{t}
\\ \text{E}(T) &= \lambda^{-1} \cdot \Gamma\left(1 + \frac{1}{p}\right)
\end{aligned}
$$
::: callout-note
Recall from calculus:
- $\Gamma(t) \stackrel{\text{def}}{=}\int_{u=0}^{\infty}u^{t-1}e^{-u}du$
- $\Gamma(t) = (t-1)!$ for integers $t \in \mathbb Z$
- It is implemented by the `gamma()` function in R.
```{r, echo = FALSE}
library(ggplot2)
ggplot() +
geom_function(fun = gamma) +
geom_point(aes(x = 1:5, y = gamma(1:5))) +
xlim(1,5) +
xlab("t") +
ylab(expression(Gamma(t))) +
theme_bw() +
theme(axis.title.y = element_text(angle=0)) +
expand_limits(y = 0)
```
:::
Here are some Weibull density functions, with $\lambda = 1$ and $p$
varying:
```{r}
#| fig-cap: "Density functions for Weibull distribution"
library(ggplot2)
lambda = 1
ggplot() +
geom_function(
aes(col = "0.25"),
fun = \(x) dweibull(x, shape = 0.25, scale = 1/lambda)) +
geom_function(
aes(col = "0.5"),
fun = \(x) dweibull(x, shape = 0.5, scale = 1/lambda)) +
geom_function(
aes(col = "1"),
fun = \(x) dweibull(x, shape = 1, scale = 1/lambda)) +
geom_function(
aes(col = "1.5"),
fun = \(x) dweibull(x, shape = 1.5, scale = 1/lambda)) +
geom_function(
aes(col = "2"),
fun = \(x) dweibull(x, shape = 2, scale = 1/lambda)) +
geom_function(
aes(col = "5"),
fun = \(x) dweibull(x, shape = 5, scale = 1/lambda)) +
theme_bw() +
xlim(0, 2.5) +
ylab("f(t)") +
theme(axis.title.y = element_text(angle=0)) +
theme(legend.position="bottom") +
guides(
col =
guide_legend(
title = "p",
label.theme =
element_text(
size = 12)))
```
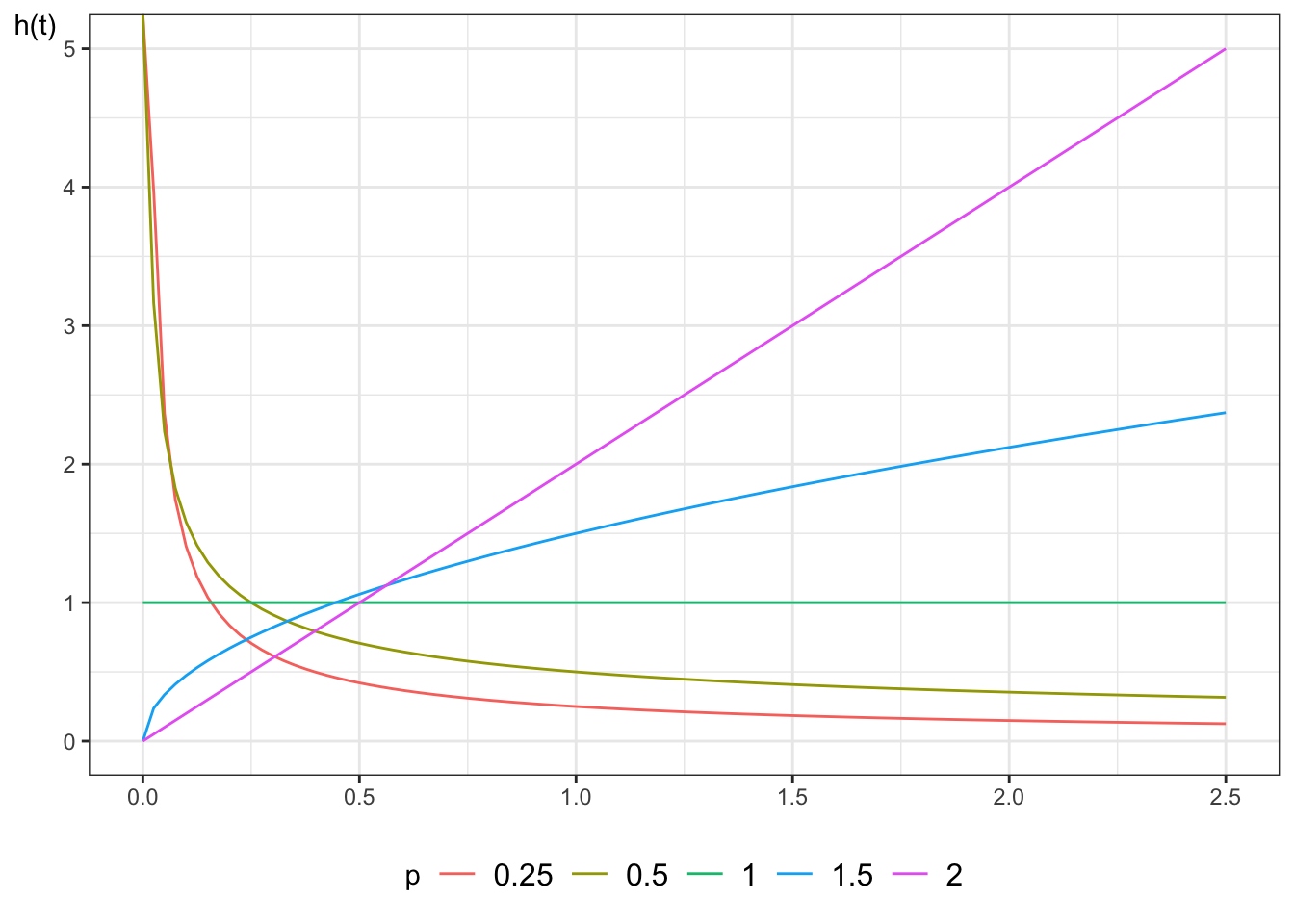
#### Properties of Weibull hazard functions
:::{#thm-weibull-props}
If $T$ has a Weibull distribution, then:
- When $p=1$, the Weibull distribution simplifies to the exponential
distribution
- When $p > 1$, the hazard is increasing: $h'(t) > 0$
- When $p < 1$, the hazard is decreasing: $h'(t) < 0$
- $\log{\cuhaz(t)}$ is a straight line relative to $\log{t}$:
$\log{\cuhaz(t)} = p \log{\lambda} + p \log{t}$
:::
---
:::{#exr-weibull}
Prove @thm-weibull-props.
:::
---
::: notes
The Weibull distribution provides more flexibility than the exponential.
@fig-exm-weibull-hazards shows some Weibull hazard functions,
with $\lambda = 1$ and $p$ varying:
:::
```{r}
#| label: fig-exm-weibull-hazards
#| fig-cap: "Hazard functions for Weibull distribution"
library(ggplot2)
library(eha)
lambda = 1
ggplot() +
geom_function(
aes(col = "0.25"),
fun = \(x) hweibull(x, shape = 0.25, scale = 1/lambda)) +
geom_function(
aes(col = "0.5"),
fun = \(x) hweibull(x, shape = 0.5, scale = 1/lambda)) +
geom_function(
aes(col = "1"),
fun = \(x) hweibull(x, shape = 1, scale = 1/lambda)) +
geom_function(
aes(col = "1.5"),
fun = \(x) hweibull(x, shape = 1.5, scale = 1/lambda)) +
geom_function(
aes(col = "2"),
fun = \(x) hweibull(x, shape = 2, scale = 1/lambda)) +
theme_bw() +
xlim(0, 2.5) +
ylab(expr(lambda)) +
theme(axis.title.y = element_text(angle=0)) +
theme(legend.position="bottom") +
guides(
col =
guide_legend(
title = "p",
label.theme =
element_text(
size = 12)))
```
---
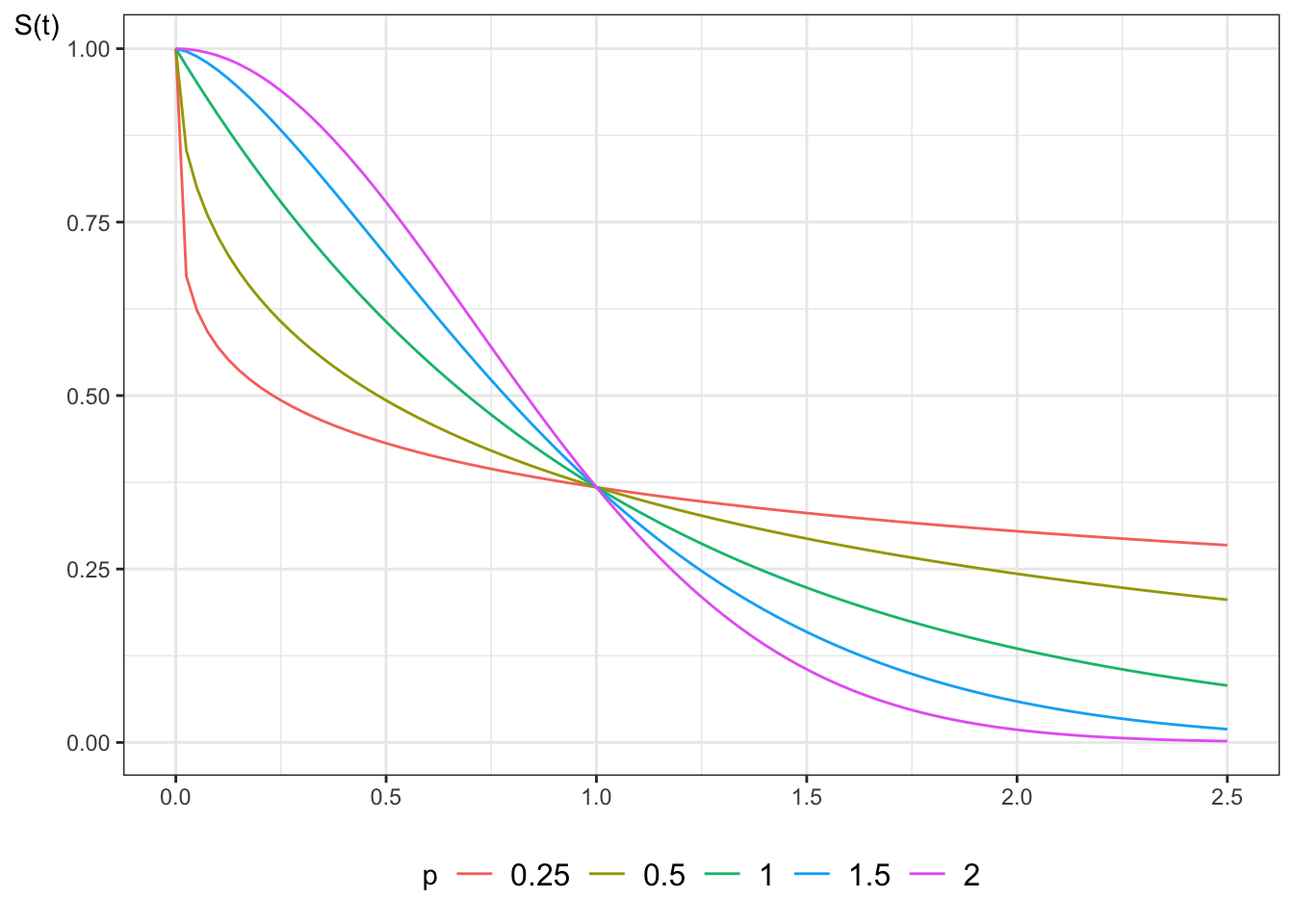
```{r}
#| label: fig-surv-weibull
#| fig-cap: "Survival functions for Weibull distribution"
library(ggplot2)
lambda = 1
ggplot() +
geom_function(
aes(col = "0.25"),
fun = \(x) pweibull(lower = FALSE, x, shape = 0.25, scale = 1/lambda)) +
geom_function(
aes(col = "0.5"),
fun = \(x) pweibull(lower = FALSE, x, shape = 0.5, scale = 1/lambda)) +
geom_function(
aes(col = "1"),
fun = \(x) pweibull(lower = FALSE, x, shape = 1, scale = 1/lambda)) +
geom_function(
aes(col = "1.5"),
fun = \(x) pweibull(lower = FALSE, x, shape = 1.5, scale = 1/lambda)) +
geom_function(
aes(col = "2"),
fun = \(x) pweibull(lower = FALSE, x, shape = 2, scale = 1/lambda)) +
theme_bw() +
xlim(0, 2.5) +
ylab("S(t)") +
theme(axis.title.y = element_text(angle=0)) +
theme(legend.position="bottom") +
guides(
col =
guide_legend(
title = "p",
label.theme =
element_text(
size = 12)))
```
### Exponential Regression
For each subject $i$, define a linear predictor:
$$
\begin{aligned}
\eta(\vec x) &= \beta_0 + (\beta_1x_1 + \dots + \beta_p x_p)\\
\haz(t|\vec x) &= \exp{\eta(\vec x)}\\
\haz_0 &\stackrel{\text{def}}{=} \haz(t|\vec 0)\\
&= \exp{\eta(\vec 0)}\\
&= \exp{\beta_0 + (\beta_1 \cdot 0 + \dots + \beta_p \cdot 0)}\\
&= \exp{\beta_0 + 0}\\
&= \exp{\beta_0}\\
\end{aligned}
$$
We let the linear predictor have a constant term, and when there are no
additional predictors the hazard is $\lambda = \exp{\beta_0}$. This has
a log link as in a generalized linear model. Since the hazard does not
depend on $t$, the hazards are (trivially) proportional.
### Accelerated Failure Time
Previously, we assumed the hazards were proportional; that is, the
covariates multiplied the baseline hazard function:
$$
\begin{aligned}
h(T=t|X=x)
&\stackrel{\text{def}}{=} p(T=t|X=x,T \ge t)\\
&= \haz(t|X=0)\cdot \exp{\eta(x)}\\
&= \haz(t|X=0)\cdot \theta(x)\\
&= \haz_0(t)\cdot \theta(x)
\end{aligned}
$$
and correspondingly,
$$
\begin{aligned}
\cuhaz(t|x)
&= \theta(x)\cuhaz_0(t)\\
\surv(t|x)
&= \expf{-\cuhaz(t|x)}\\
&= \expf{-\theta(x)\cdot \cuhaz_0(t)}\\
&= \paren{\expf{- \cuhaz_0(t)}}^{\theta(x)}\\
&= \paren{\surv_0(t)}^{\theta(x)}\\
\end{aligned}
$$
An alternative modeling assumption would be
$$\surv(t|X=x)=\surv_0(t\cdot \theta(x))$$ where $\theta(x)=\expf{\eta(x)}$,
$\eta(x) =\beta_1x_1+\cdots+\beta_px_p$, and $\surv_0(t)=\P(T\ge t|X=0)$ is
the base survival function.
Then
$$
\begin{aligned}
\Expp[T|X=x]
&= \int_{t=0}^{\infty} \surv(t|x)dt\\
&= \int_{t=0}^{\infty} \surv_0(t\cdot \theta(x))dt\\
&= \int_{u=0}^{\infty} \surv_0(u)du \cdot \theta(x)^{-1}\\
&= \theta(x)^{-1} \cdot \int_{u=0}^{\infty} \surv_0(u)du\\
&= \theta(x)^{-1} \cdot \Expp[T|X=0]\\
\end{aligned}
$$ So the mean of $T$ given $X=x$ is the baseline mean divided by
$\theta(x) = \exp{\eta(x)}$.
This modeling strategy is called an accelerated failure time model,
because covariates cause uniform acceleration (or slowing) of failure
times.
Additionally:
$$
\begin{aligned}
\cuhaz(t|x) &= \cuhaz_0(\theta(x)\cdot t)\\
\haz(t|x) &= \theta(x) \cdot \haz_0(\theta(x)\cdot t)
\end{aligned}
$$
If the base distribution is exponential with parameter $\lambda$ then
$$
\begin{aligned}
\surv(t|x)
&= \exp{-\lambda \cdot t \theta(x)}\\
&= [\exp{-\lambda t}]^{\theta(x)}\\
\end{aligned}
$$
which is an exponential model with base hazard multiplied by
$\theta(x)$, which is also the proportional hazards model.
::: hidden
In terms of the log survival time $Y=\log{T}$ the model can be written
as
$$
\begin{aligned}
Y&=\alpha-\eta+W\\
\alpha&= -\log{\lambda}
\end{aligned}
$$
where $W$ has the extreme value distribution. The estimated parameter
$\lambda$ is the intercept and the other coefficients are those of
$\eta$, which will be the opposite sign of those for coxph.
:::
For a Weibull distribution, the hazard function and the survival
function are
$$
\begin{aligned}
\haz(t)&=\lambda p (\lambda t)^{p-1}\\
\surv(t)&=e^{-(\lambda t)^p}
\end{aligned}
$$
We can construct a proportional hazards model by using a linear
predictor $\eta_i$ without constant term and letting
$\theta_i=e^{\eta_i}$ we have
$$
\begin{aligned}
\haz(t)&=\lambda p (\lambda t)^{p-1}\theta_i
\end{aligned}
$$
A distribution with $\haz(t)=\lambda p (\lambda t)^{p-1}\theta_i$ is a
Weibull distribution with parameters $\lambda^*=\lambda \theta_i^{1/p}$
and $p$ so the survival function is
$$
\begin{aligned}
S^*(t)&=e^{-(\lambda^* t)^p}\\
&=e^{-(\lambda \theta^{1/p} t)^p}\\
&= \surv(t\theta^{1/p})
\end{aligned}
$$
so this is also an accelerated failure time model.
::: hidden
In terms of the log survival time $Y=\log{T}$ the model can be written
as
$$
\begin{aligned}
Y&=\alpha-\sigma\eta+\sigma W\\
\alpha&= -\log{\lambda}\\
\sigma &= 1/p
\end{aligned}
$$
where $W$ has the extreme value distribution. The estimated parameter
$\lambda$ is the intercept and the other coefficients are those of
$\eta$, which will be the opposite sign of those for `coxph`.
:::
These AFT models are log-linear, meaning that the linear predictor has a
log link. The exponential and the Weibull are the only log-linear models
that are simultaneously proportional hazards models. Other parametric
distributions can be used for survival regression either as a
proportional hazards model or as an accelerated failure time model.
### Dataset: Leukemia treatments
Remission survival times on 42 leukemia patients, half on new treatment,
half on standard treatment.
This is the same data as the `drug6mp` data from KMsurv, but with two
other variables and without the pairing.
```{r}
#| eval: false
library(haven)
library(survival)
anderson =
paste0(
"http://web1.sph.emory.edu/dkleinb/allDatasets",
"/surv2datasets/anderson.dta") |>
read_dta() |>
mutate(
status = status |>
case_match(
1 ~ "relapse",
0 ~ "censored"
),
sex = sex |>
case_match(
0 ~ "female",
1 ~ "male"
),
rx = rx |>
case_match(
0 ~ "new",
1 ~ "standard"
),
surv = Surv(time = survt,event = (status == "relapse"))
)
print(anderson)
```
```{r}
#| include: false
#| label: anderson-load-local
library(haven)
library(survival)
anderson =
fs::path_package(
"rme", "extdata/anderson.dta") |>
read_dta() |>
mutate(
status = status |>
case_match(
1 ~ "relapse",
0 ~ "censored"
),
sex = sex |>
case_match(
0 ~ "female",
1 ~ "male"
),
rx = rx |>
case_match(
0 ~ "new",
1 ~ "standard"
),
surv = Surv(time = survt,event = (status == "relapse"))
)
print(anderson)
```
#### Cox semi-parametric model
```{r}
anderson.cox0 = coxph(
formula = surv ~ rx,
data = anderson)
summary(anderson.cox0)
```
#### Weibull parametric model
```{r}
anderson.weib <- survreg(
formula = surv ~ rx,
data = anderson,
dist = "weibull")
summary(anderson.weib)
```
#### Exponential parametric model
```{r}
anderson.exp <- survreg(
formula = surv ~ rx,
data = anderson,
dist = "exp")
summary(anderson.exp)
```
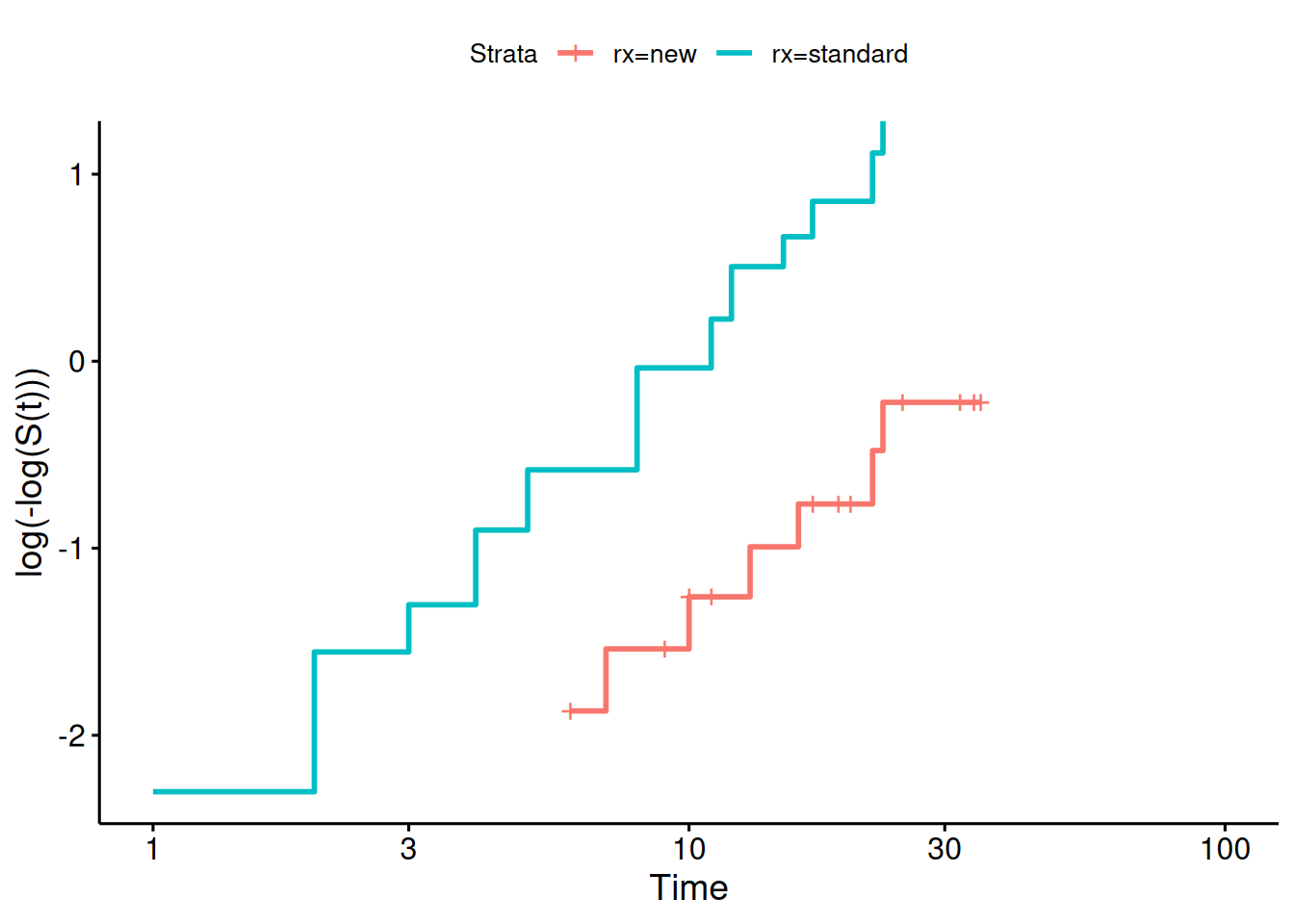
#### Diagnostic - complementary log-log survival plot
```{r}
library(survminer)
survfit(
formula = surv ~ rx,
data = anderson) |>
ggsurvplot(fun = "cloglog")
```
If the cloglog plot is linear, then a Weibull model may be ok.
## Combining left-truncation and interval-censoring
From [https://stat.ethz.ch/pipermail/r-help/2015-August/431733.html]:
> coxph does left truncation but not left (or interval) censoring
> survreg does interval censoring but not left truncation (or time dependent covariates).
- Terry Therneau, August 31, 2015