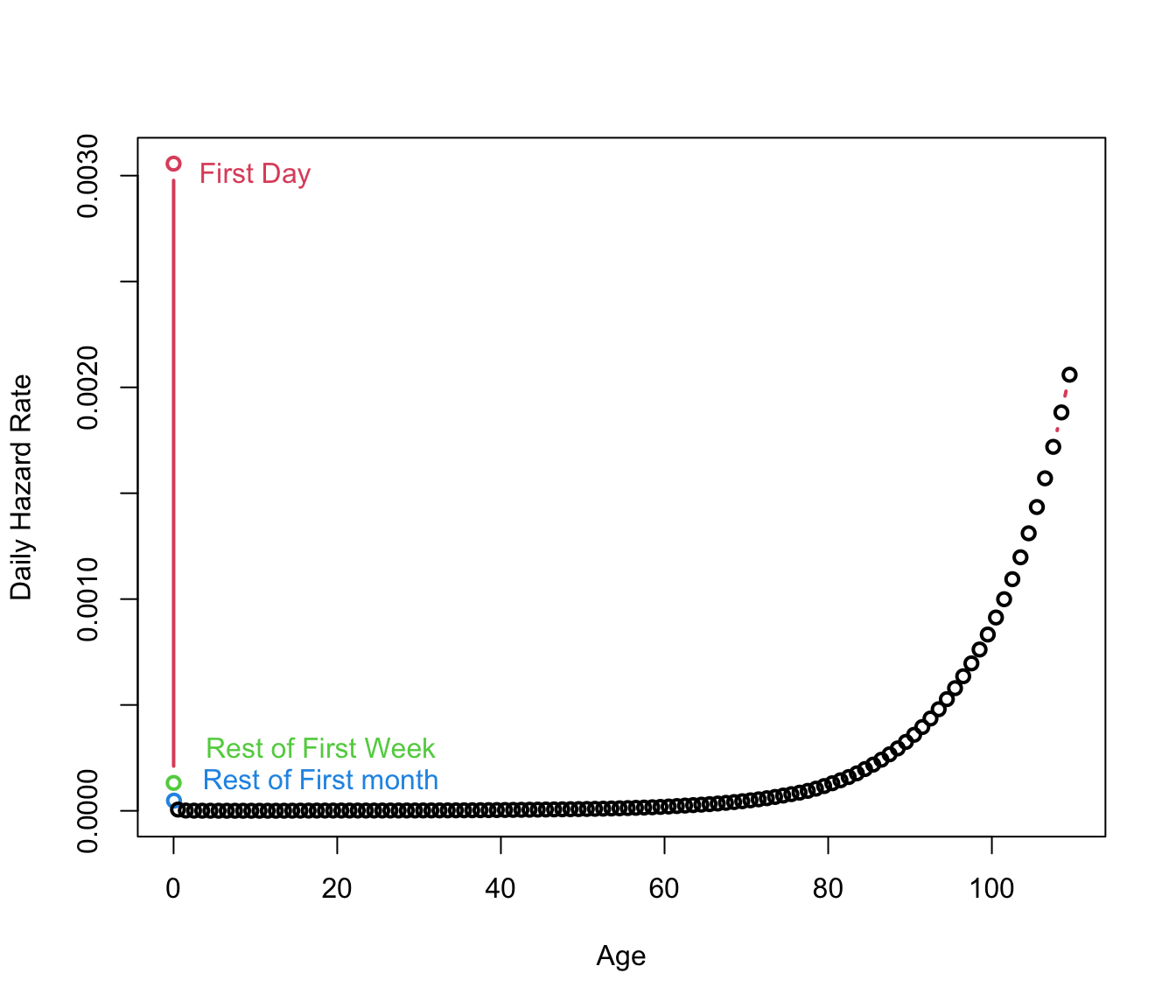
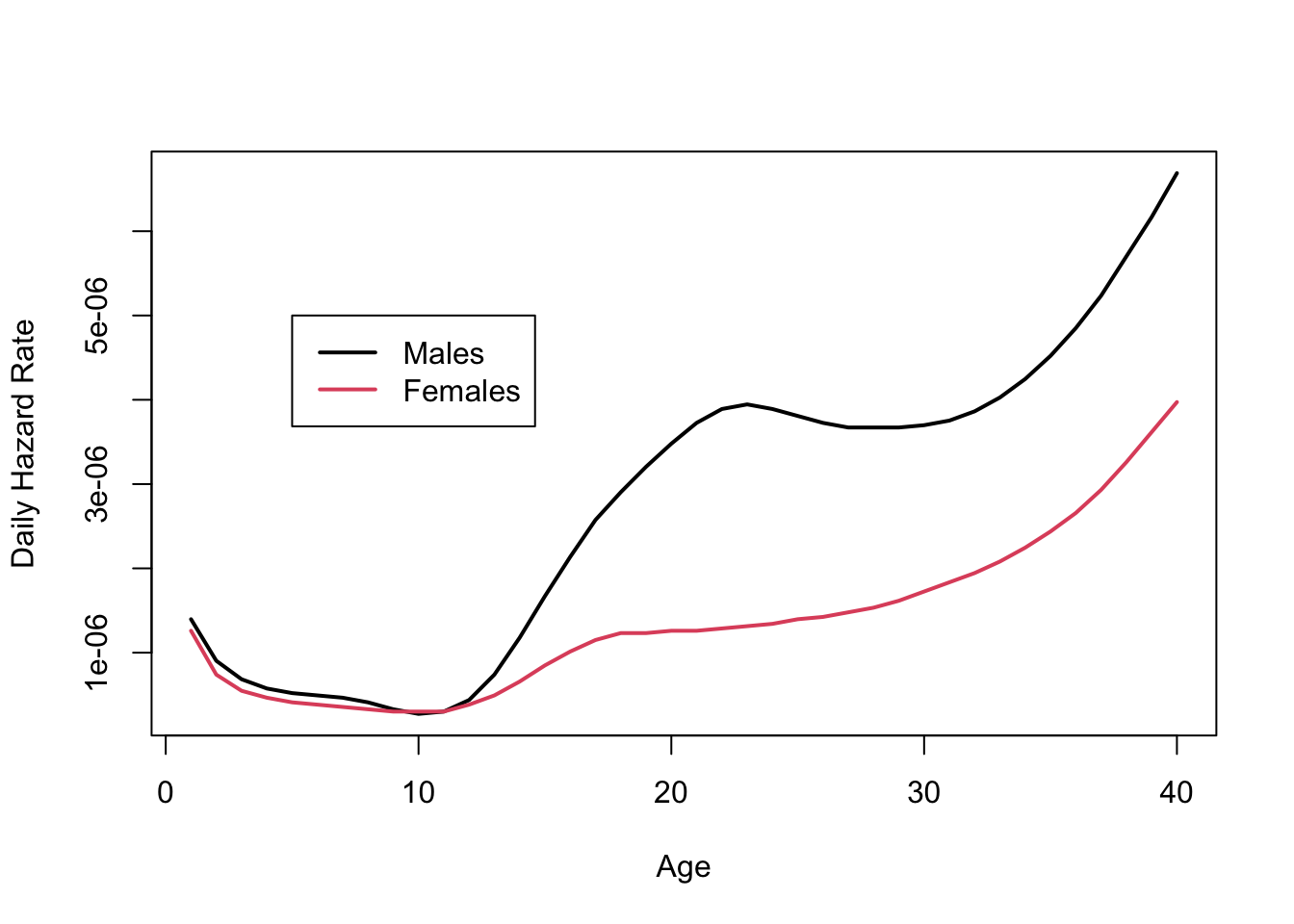
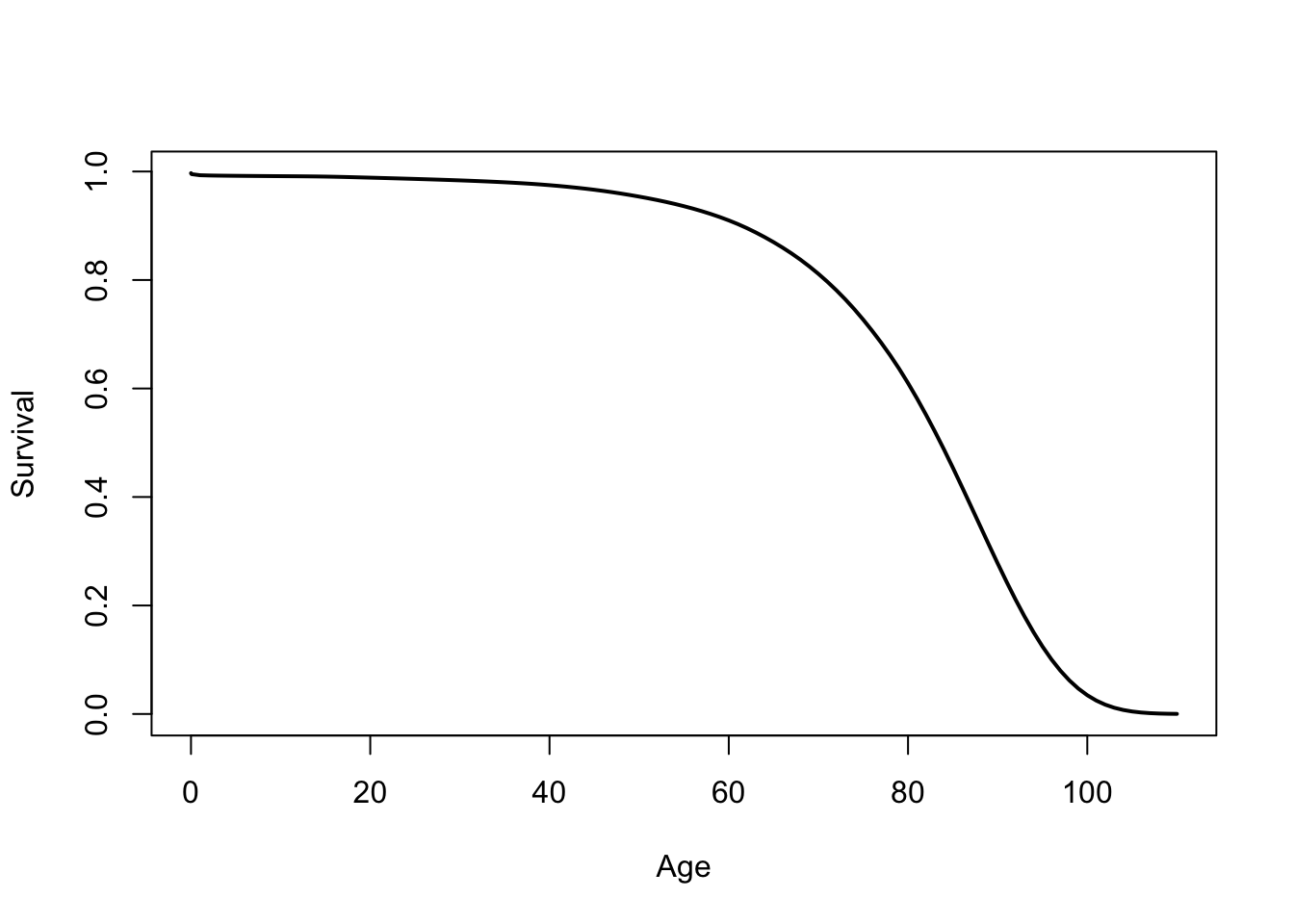
The Hazard Function
Another important quantity is the hazard function:
Definition 6.2 (Hazard function, hazard rate, hazard rate function)
The hazard function, hazard rate, hazard rate function, for a random variable \(T\) at value \(t\), typically denoted as \(\text{h}(t)\) or \(\lambda(t)\), is the conditional density of \(T\) at \(t\), given \(T \ge t\). That is:
\[{\lambda}(t) \stackrel{\text{def}}{=}\text{p}(T=t|T\ge t)\]
If \(T\) represents the time at which an event occurs, then \({\lambda}(t)\) is the probability that the event occurs at time \(t\), given that it has not occurred prior to time \(t\).
Definition 6.3 (Incidence rate) Given a population of \(N\) individuals indexed by \(i\), each with their own hazard rate \({\lambda}_i(t)\), the incidence rate for that population is the mean hazard rate:
\[\bar{{\lambda}}(t) \stackrel{\text{def}}{=}\frac{1}{N}\sum_{i=1}^N{\lambda}_i(t)\]
Theorem 6.4 (Incidence rate in a homogenous population) If a population of individuals indexed by \(i\) all have identical hazard rates \({\lambda}_i(t) = {\lambda}(t)\), then the incidence rate for that population is equal to the hazard rate:
\[\bar{{\lambda}}(t) = {\lambda}(t)\]
The hazard function has an important relationship to the density and survival functions, which we can use to derive the hazard function for a given probability distribution (Theorem 6.5).
Lemma 6.1 (Joint probability of a variable with itself) \[p(T=t, T\ge t) = p(T=t)\]
Proof. Recall from Epi 202: if \(A\) and \(B\) are statistical events and \(A\subseteq B\), then \(p(A, B) = p(A)\). In particular, \(\{T=t\} \subseteq \{T\geq t\}\), so \(p(T=t, T\ge t) = p(T=t)\).
Theorem 6.5 (Hazard equals density over survival) \[{\lambda}(t)=\frac{f(t)}{\text{S}(t)}\]
Proof. \[
\begin{aligned}
{\lambda}(t) &=p(T=t|T\ge t)\\
&=\frac{p(T=t, T\ge t)}{p(T \ge t)}\\
&=\frac{p(T=t)}{p(T \ge t)}\\
&=\frac{f(t)}{\text{S}(t)}
\end{aligned}
\]
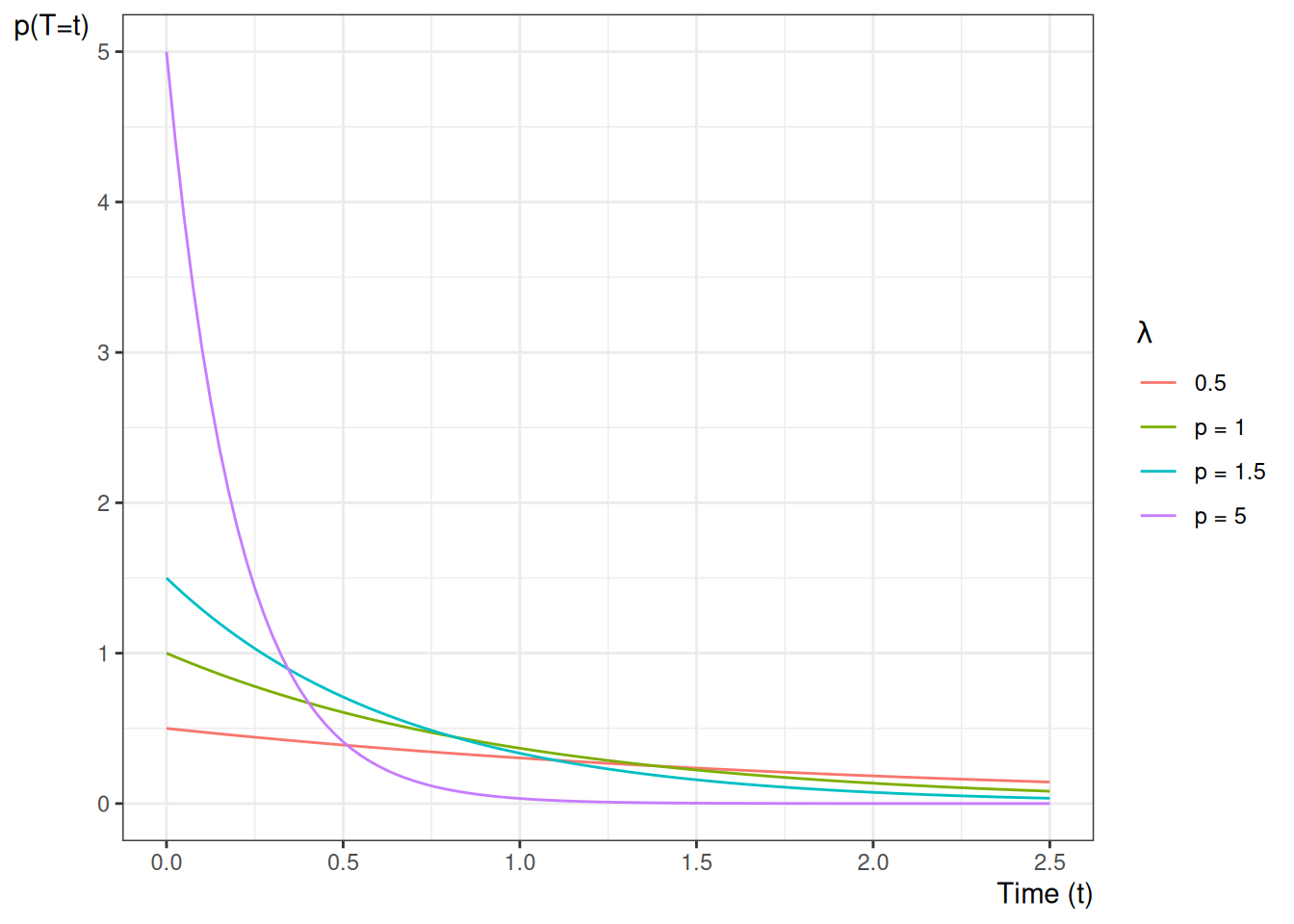
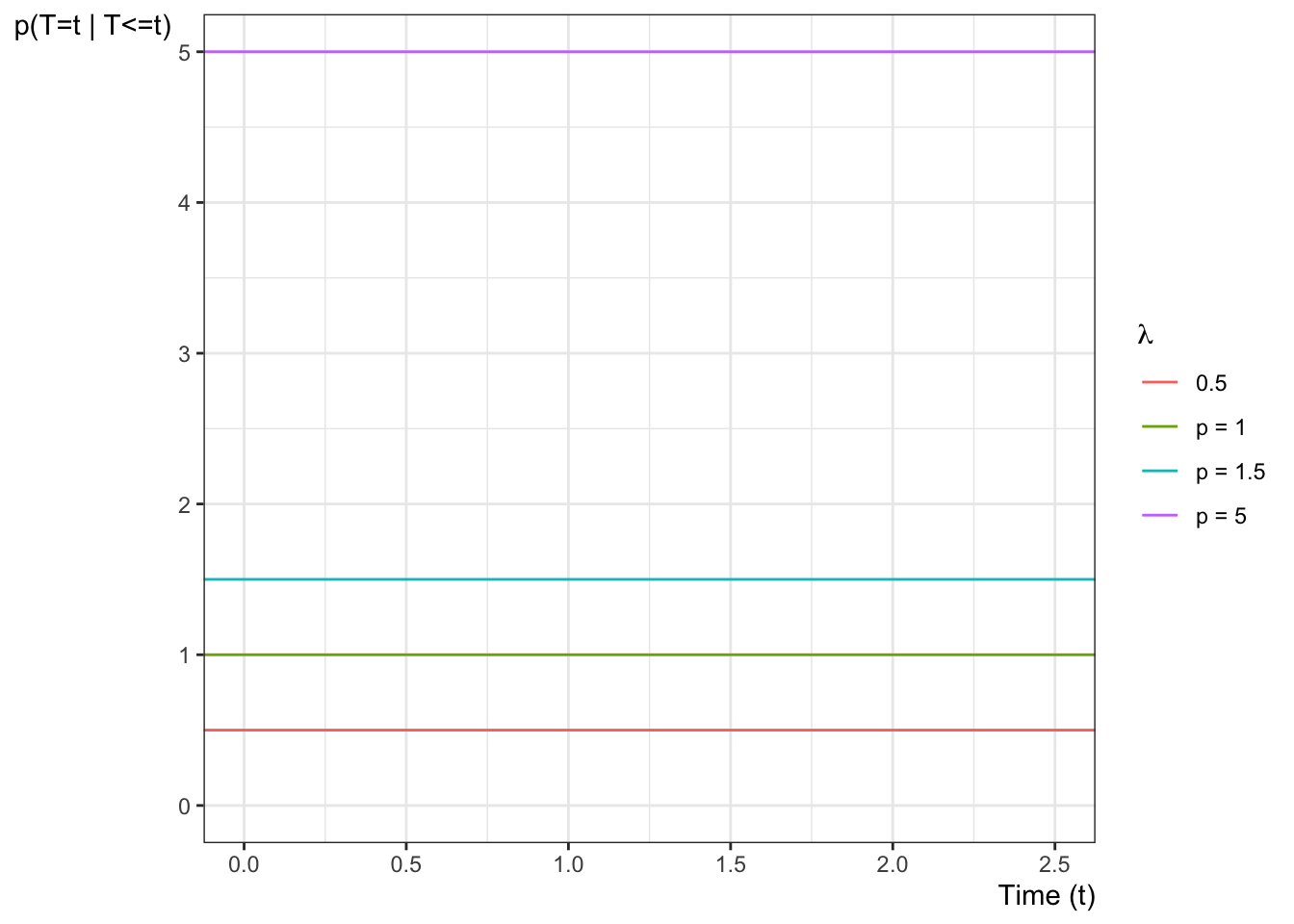
Example 6.4 (exponential distribution) The hazard function of the exponential distribution family of models is:
\[
\begin{aligned}
P(T=t|T \ge t)
&= \frac{f(t)}{\text{S}(t)}\\
&= \frac{\mathbb{1}_{t \ge 0}\cdot \lambda \text{e}^{-\lambda t}}{\text{e}^{-\lambda t}}\\
&=\mathbb{1}_{t \ge 0}\cdot \lambda
\end{aligned}
\] Figure 6.2 shows some examples of exponential hazard functions.
We can also view the hazard function as the derivative of the negative of the logarithm of the survival function:
Theorem 6.6 (transform survival to hazard) \[{\lambda}(t) = \frac{\partial}{\partial t}{\left\{-\log{\text{S}(t)}\right\}}\]
Proof. \[
\begin{aligned}
{\lambda}(t)
&= \frac{f(t)}{\text{S}(t)}\\
&= \frac{-S'(t)}{\text{S}(t)}\\
&= -\frac{S'(t)}{\text{S}(t)}\\
&=-\frac{\partial}{\partial t}\log{\text{S}(t)}\\
&=\frac{\partial}{\partial t}{\left\{-\log{\text{S}(t)}\right\}}
\end{aligned}
\]
Definition 6.4 (hazard ratio) \[\theta(t| \tilde{x}: {\tilde{x}^*}) \stackrel{\text{def}}{=}\frac{{\lambda}(t|\tilde{x})}{{\lambda}(t|{\tilde{x}^*})}\]
The Cumulative Hazard Function
Since \({\lambda}(t) = \frac{\partial}{\partial t}{\left\{-\log{\text{S}(t)}\right\}}\) (see Theorem 6.6), we also have:
Corollary 6.1 \[\text{S}(t) = \text{exp}{\left\{-\int_{u=0}^t {\lambda}(u)du\right\}} \tag{6.1}\]
The integral in Equation 6.1 is important enough to have its own name: cumulative hazard.
Definition 6.5 (cumulative hazard) The cumulative hazard function, often denoted \(\Lambda(t)\) or \(\text{H}(t)\), is defined as:
\[{\Lambda}(t) \stackrel{\text{def}}{=}\int_{u=0}^t {\lambda}(u) du\]
As we will see below, \({\Lambda}(t)\) is tractable to estimate, and we can then derive an estimate of the hazard function using an approximate derivative of the estimated cumulative hazard.
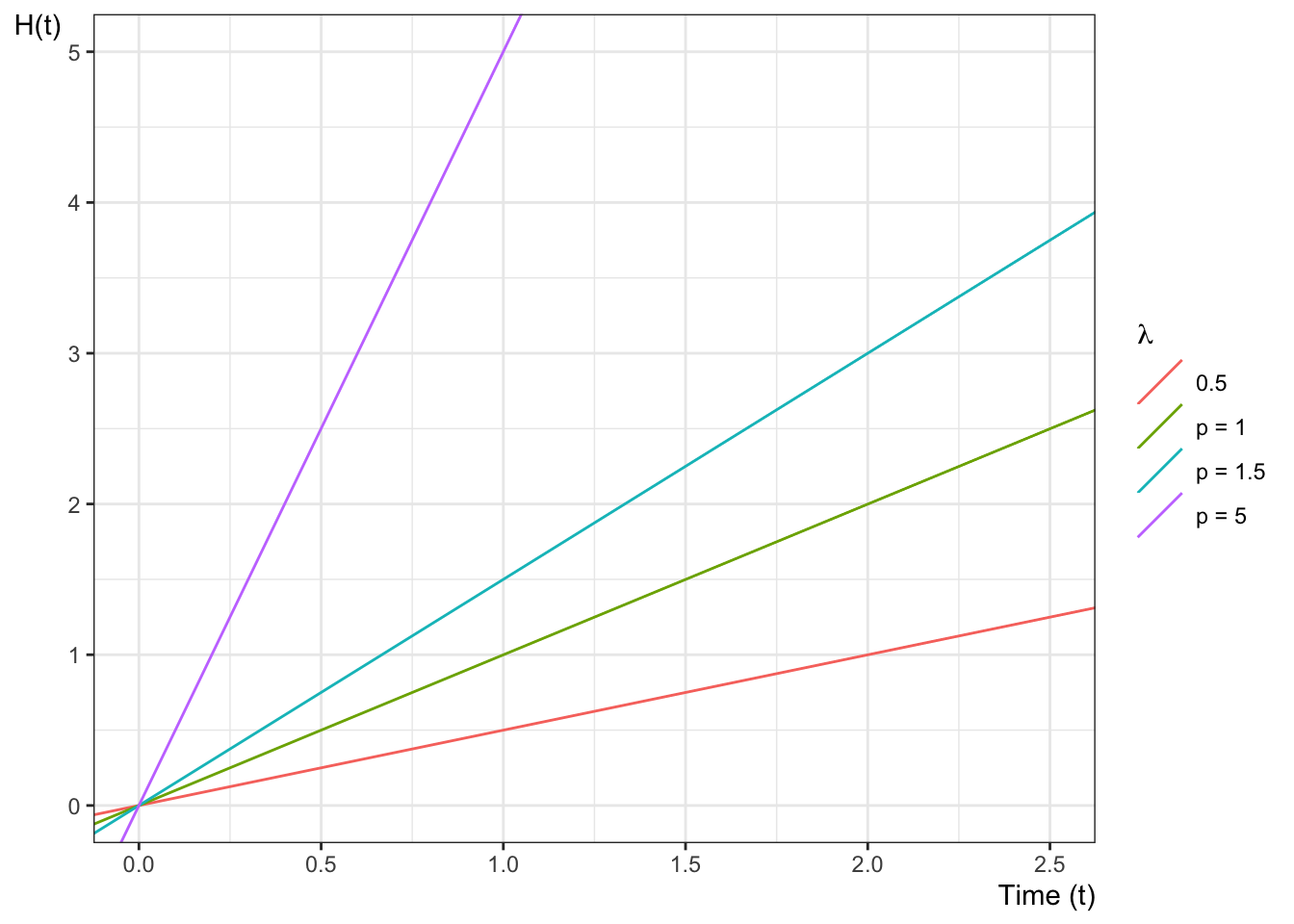
Example 6.5 The cumulative hazard function for the exponential distribution with rate parameter \(\lambda\) is:
\[
{\Lambda}(t) = \mathbb{1}_{t \ge 0}\cdot \lambda t
\]
Figure 6.3 shows some examples of exponential cumulative hazard functions.
Likelihood with censoring
If an event time \(T\) is observed exactly as \(T=t\), then the likelihood of that observation is just its probability density function:
\[
\begin{aligned}
\mathscr{L}(t)
&= \text{f}(T=t)\\
&\stackrel{\text{def}}{=}\text{f}_T(t)\\
&= {\lambda}_T(t)\text{S}_T(t)\\
\ell(t)
&\stackrel{\text{def}}{=}\text{log}{\left\{\mathscr{L}(t)\right\}}\\
&= \text{log}{\left\{{\lambda}_T(t) \text{S}_T(t)\right\}}\\
&= \text{log}{\left\{{\lambda}_T(t)\right\}} + \text{log}{\left\{\text{S}_T(t)\right\}}\\
&= \text{log}{\left\{{\lambda}_T(t)\right\}} - {\Lambda}_T(t)\\
\end{aligned}
\]
If instead the event time \(T\) is censored and only known to be after time \(y\), then the likelihood of that censored observation is instead the survival function evaluated at the censoring time:
\[
\begin{aligned}
\mathscr{L}(y)
&=p_T(T>y)\\
&\stackrel{\text{def}}{=}\text{S}_T(y)\\
\ell(y)
&\stackrel{\text{def}}{=}\text{log}{\left\{\mathscr{L}(y)\right\}}\\
&=\text{log}{\left\{\text{S}(y)\right\}}\\
&=-{\Lambda}(y)\\
\end{aligned}
\]
What’s written above is incomplete. We also observed whether or not the observation was censored. Let \(C\) denote the time when censoring would occur (if the event did not occur first); let \(f_C(y)\) and \(S_C(y)\) be the corresponding density and survival functions for the censoring event.
Let \(Y\) denote the time when observation ended (either by censoring or by the event of interest occurring), and let \(D\) be an indicator variable for the event occurring at \(Y\) (so \(D=0\) represents a censored observation and \(D=1\) represents an uncensored observation). In other words, let \(Y \stackrel{\text{def}}{=}\min(T,C)\) and \(D \stackrel{\text{def}}{=}\mathbb 1{\{T<=C\}}\).
Then the complete likelihood of the observed data \((Y,D)\) is:
\[
\begin{aligned}
\mathscr{L}(y,d)
&= \text{p}(Y=y, D=d)\\
&= {\left[\text{p}(T=y,C> y)\right]}^d \cdot {\left[\text{p}(T>y,C=y)\right]}^{1-d}\\
\end{aligned}
\]
Typically, survival analyses assume that \(C\) and \(T\) are mutually independent; this assumption is called “non-informative” censoring.
Then the joint likelihood \(\text{p}(Y,D)\) factors into the product \(\text{p}(Y), \text{p}(D)\), and the likelihood reduces to:
\[
\begin{aligned}
\mathscr{L}(y,d)
&= {\left[\text{p}(T=y,C> y)\right]}^d\cdot
{\left[\text{p}(T>y,C=y)\right]}^{1-d}\\
&= {\left[\text{p}(T=y)\text{p}(C> y)\right]}^d\cdot
{\left[\text{p}(T>y)\text{p}(C=y)\right]}^{1-d}\\
&= {\left[\text{f}_T(y)\text{S}_C(y)\right]}^d\cdot
{\left[\text{S}(y)\text{f}_C(y)\right]}^{1-d}\\
&= {\left[\text{f}_T(y)^d \text{S}_C(y)^d\right]}\cdot
{\left[\text{S}_T(y)^{1-d} \text{f}_C(y)^{1-d}\right]}\\
&= {\left(\text{f}_T(y)^d \cdot \text{S}_T(y)^{1-d}\right)}\cdot
{\left(\text{f}_C(y)^{1-d} \cdot \text{S}_C(y)^{d}\right)}
\end{aligned}
\]
The corresponding log-likelihood is:
\[
\begin{aligned}
\ell(y,d)
&= \text{log}{\left\{\mathscr{L}(y,d)\right\}}\\
&= \text{log}{\left\{
{\left(f_T(y)^d \cdot S_T(y)^{1-d}\right)}
\cdot
{\left(f_C(y)^{1-d} \cdot S_C(y)^{d}\right)}
\right\}}
\\
&= \text{log}{\left\{f_T(y)^d \cdot S_T(y)^{1-d}\right\}}
+
\text{log}{\left\{f_C(y)^{1-d} \cdot S_C(y)^{d}\right\}}
\end{aligned}
\] Let
-
\(\theta_T\) represent the parameters of \(p_T(t)\),
-
\(\theta_C\) represent the parameters of \(p_C(c)\),
-
\(\theta = (\theta_T, \theta_C)\) be the combined vector of all parameters.
The corresponding score function is:
\[
\begin{aligned}
\ell'(y,d)
&= \frac{\partial}{\partial \theta}
{\left[
\text{log}{\left\{f_T(y)^d \cdot S_T(y)^{1-d}\right\}}
+
\text{log}{\left\{f_C(y)^{1-d} \cdot S_C(y)^{d}\right\}}
\right]}
\\
&=
{\left(
\frac{\partial}{\partial \theta}
\text{log}{\left\{
f_T(y)^d \cdot S_T(y)^{1-d}
\right\}}
\right)}
+
{\left(
\frac{\partial}{\partial \theta}
\text{log}{\left\{
f_C(y)^{1-d} \cdot S_C(y)^{d}
\right\}}
\right)}
\end{aligned}
\]
As long as \(\theta_C\) and \(\theta_T\) don’t share any parameters, then if censoring is non-informative, the partial derivative with respect to \(\theta_T\) is:
\[
\begin{aligned}
\ell'_{\theta_T}(y,d)
&\stackrel{\text{def}}{=}\frac{\partial}{\partial \theta_T}\ell(y,d)\\
&=
\left(
\frac{\partial}{\partial \theta_T}
\text{log}\left\{
f_T(y)^d \cdot S_T(y)^{1-d}
\right\}
\right)
+
\left(
\frac{\partial}{\partial \theta_T}
\text{log}\left\{
f_C(y)^{1-d} \cdot S_C(y)^{d}
\right\}
\right)\\
&=
\left(
\frac{\partial}{\partial \theta_T}
\text{log}\left\{
f_T(y)^d \cdot S_T(y)^{1-d}
\right\}
\right) + 0\\
&=
\frac{\partial}{\partial \theta_T}
\text{log}\left\{
f_T(y)^d \cdot S_T(y)^{1-d}
\right\}\\
\end{aligned}
\]
Thus, the MLE for \(\theta_T\) won’t depend on \(\theta_C\), and we can ignore the distribution of \(C\) when estimating the parameters of \(f_T(t)=p(T=t)\).
Then:
\[
\begin{aligned}
\mathscr{L}(y,d)
&= f_T(y)^d \cdot S_T(y)^{1-d}\\
&= \left(h_T(y)^d S_T(y)^d\right) \cdot S_T(y)^{1-d}\\
&= h_T(y)^d \cdot S_T(y)^d \cdot S_T(y)^{1-d}\\
&= h_T(y)^d \cdot S_T(y)\\
&= S_T(y) \cdot h_T(y)^d \\
\end{aligned}
\]
That is, if the event occurred at time \(y\) (i.e., if \(d=1\)), then the likelihood of \((Y,D) = (y,d)\) is equal to the hazard function at \(y\) times the survival function at \(y\). Otherwise, the likelihood is equal to just the survival function at \(y\).
The corresponding log-likelihood is:
\[
\begin{aligned}
\ell(y,d)
&=\text{log}\left\{\mathscr{L}(y,d)\right\}\\
&= \text{log}\left\{S_T(y) \cdot h_T(y)^d\right\}\\
&= \text{log}\left\{S_T(y)\right\} + \text{log}\left\{h_T(y)^d\right\}\\
&= \text{log}\left\{S_T(y)\right\} + d\cdot \text{log}\left\{h_T(y)\right\}\\
&= -H_T(y) + d\cdot \text{log}\left\{h_T(y)\right\}\\
\end{aligned}
\]
In other words, the log-likelihood contribution from a single observation \((Y,D) = (y,d)\) is equal to the negative cumulative hazard at \(y\), plus the log of the hazard at \(y\) if the event occurred at time \(y\).